Update: This paper has now been published at eLife.
“Tracking the popularity and outcomes of all bioRxiv preprints” by Richard J. Abdill and Ran Blekhman
https://www.biorxiv.org/content/10.1101/515643v1
In this article, Abdill and Blekhman describe a database of bioRxiv preprints and associated data, present various analyses, and introduce a website, Rxivist.org, for sorting bioRxiv preprints based on Twitter activity and PDF downloads.
I selected this article for two reasons. First, the analysis received a lot of attention on social media, particularly (1) correlations between Journal Impact Factor and preprint popularity, (2) delays between preprint and journal publication and (3) the fraction of preprints that are eventually published in journals. Interestingly, the last point was cited as a reason preprints are unreliable, even though the Rxivist preprint itself has not been published in a peer-reviewed journal. Second, the Rxivist site provides a new way to discover preprints that could influence which preprints are read and who reads them. Discovering life sciences preprints will become more important if preprinting frequency continues to grow rapidly.
I thank the authors for sharing their work on bioRxiv and also for providing the underlying data.
I solicited reviewers from 2 scientists with expertise in science publication, Dr. Daniel Himmelstein and Dr. Devang Mehta. I thank the reviewers for taking the time to provide thorough reviews.
UPDATE 4 March 2019: Dr. Naomi Penfold (ASAPbio) brought my attention to interesting analysis on the data from this preprint by Dr. Bianca Kramer (Utrecht Library). Dr. Kramer investigated whether the relationship between bioRxiv downloads and “Impact Factor” could be explained by differences in bioRxiv downloads before and after journal publication, or by whether or not a journal was open access. Dr. Kramer’s analysis is available here.
Reviewer 1
This study (version 1) provides a thorough analysis and report on the bioRxiv preprint server from its launch in 2013 through November, 2018. The study quantifies preprint adoption in the biological sciences, analyzing specifically trends in preprint postings, downloads, authorship, and subsequent journal publication.
To facilitate the study, the authors developed a database of bioRxiv preprints. This database, which is designed to continuously update, also powers a website with preprint recommendations and an API to enable programmatic data access. If the database is continually maintained and updated, it could help substitute for the lack of any official API or bulk database dump by bioRxiv.
Overall, the study includes many of the most pressing and noteworthy analyses possible given available data. It is clearly written and thorough, and follows many best practices for open science. There is room for improvement, as I’ll describe, but the core findings are solid, and the accompanying database, web resources, and software will likely become helpful community resources.
Feedback is broken down by subsection.
Preprint submissions
The authors illustrate exponential growth in the number of bioRxiv preprints posted over time (Figure 1). One interesting finding is that posting in the Neuroscience category have grown at an astonishing rate, starting in mid 2016, outpacing growth in other popular categories such as Bioinformatics. One question that this section does not adequately address is the percentage of all biomedical publications that are posted as preprints. It would be helpful, for example, to compare the number of bioRxiv postings to new articles added to PubMed each year. Similarly, the publications-per-journal analysis in a later section could be contextualized by assessing the total number of articles published by each journal.
Preprint authors
Consolidating author strings from bioRxiv preprints into author entities is a challenging undertaking, with some amount imperfection essentially guaranteed. The authors describe the difficulties and perform a reasonable approach to consolidating authors. Their database will be helpful in allowing other researchers to avoid repeating the laborious standardization process. Without seeing a searchable/sortable list of all authors, such the one in my licensing blog post, it is hard to evaluate how effective the authors were at consolidating authors. I was able to find my own author profile on Rxivist, but didn’t see an option to search for authors.
One interesting finding from this section is that three-quarters of preprint authors are only listed on a single preprint. Therefore, there are many researchers with exactly one preprint experience (and likely not as the submitting author). This finding may be helpful when designing outreach activities related to preprinting.
Publication outcomes
Figure 3a shows that the percent of bioRxiv preprints that continue onto journal publication is around 70%, a similar percentage as was previously found for arXiv preprints. Many researchers view journal publication as an important stamp of approval and thus may reconsider their opinion of preprints given that the majority continue onto journal publication.
Another interesting finding reported in the Methods — which also points to the satisfactory quality & rigor of existing preprints — was that:
A search for 1,345 journals based on the list compiled by Stop Predatory Journals showed that bioRxiv lists zero papers appearing in those publications.
I think one explanation is that early preprint adopters were generally forward-thinking, competent, perceptive, and attuned researchers. Such researchers are unlikely to publish in predatory journals. However, another possibility is that bioRxiv is not detecting publications in predatory journals, as described in my following comment.
The section does not touch on the accuracy of bioRxiv’s “now published in” notices. While I think it’s rather safe to assume that when the notices exist, they are highly likely to be correct, it seems like it is possible some preprints have gone onto publication that is not detected by bioRxiv. It would be useful to assess this by selecting 100 or so unpublished preprints that are at least two years old and then manually investigating whether journal (or other) publication has actually occurred. It would also be sufficient to reference any works that have performed this analysis. As far as I understand, bioRxiv’s journal publication associations are automatically detected using text match of the title/authors in the Crossref database (related tweet), making it likely for there to be underreporting. Diligent authors can also email bioRxiv with notice of journal publication.
When matching “now published in” DOIs with journals, the authors performed a manual approach based on evaluating textual journal names. I would think using the journal ISSNs reported in the Crossref metadata would be a more robust, efficient, and automated approach.
This study retrieves the date of journal publication from Crossref. One issue that we’ve found when working with Crossref-derived publication dates is that many articles with a missing day-of-publication are coded as being published on the 1st of the month. Similarly, many articles with missing month-of-publication are coded as being published on January 1st. Therefore, articles Crossref marks as published on January 1 can often just indicate the article was published anytime that year. Articles marked as being published on the first of the month can often just indicate the article was published anytime that month. I ended up taking a look at the publication dates in Rxivist and Crossref and found that when examining the entire Crossref catalog, there was an enrichment of dates on the first of the year and each month. However, the distribution of month-day publication dates for preprinted journal articles was more uniform:
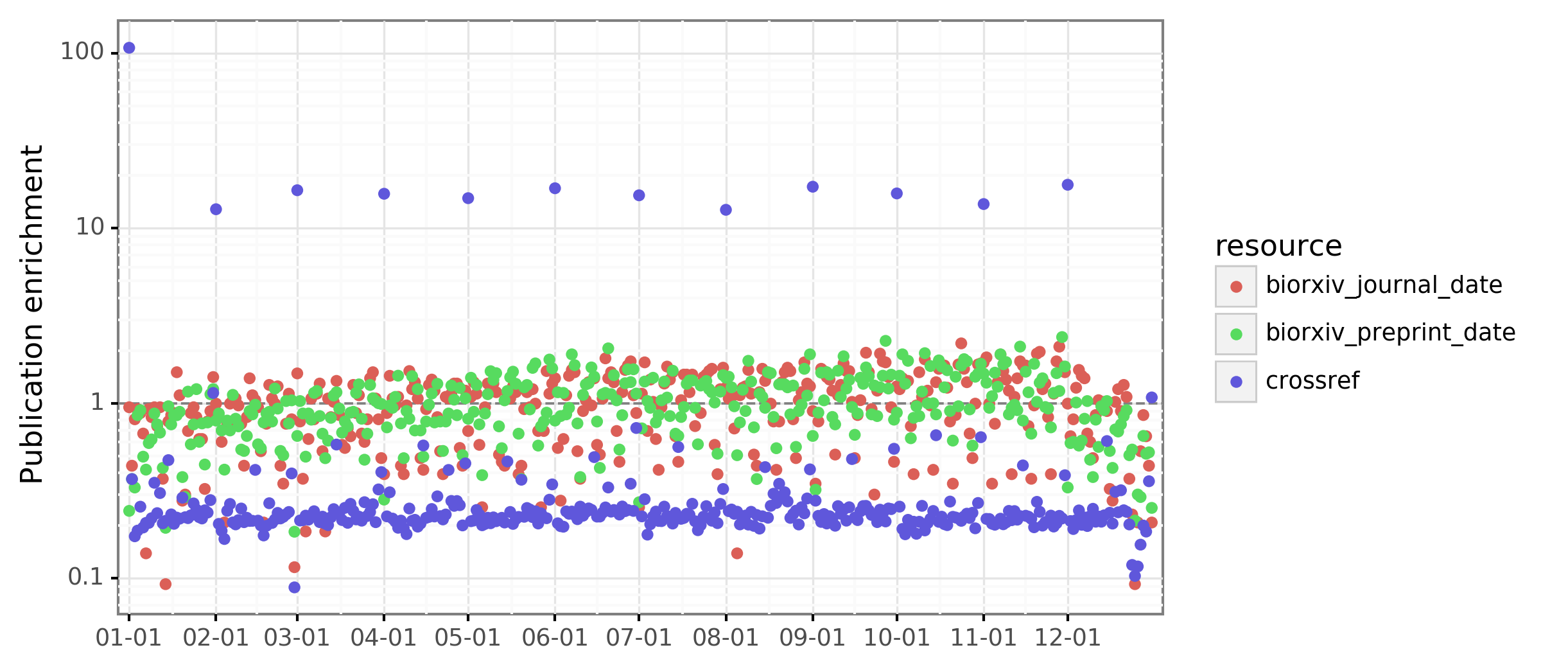
I suspect many journals have now fixed their lack of day precision. Therefore, the issue concerns mostly specific journals from the past, which are not highly represented in the bioRxiv “now published in” corpus.
Regarding the analysis of downloads versus Journal Impact Factor (JIF), I would suggest not reporting JIFs to three decimals (i.e. rather than “impact score 26.919” write “Impact Factor of 26.9), as that is false precision. The authors might also consider using “Impact Factor” rather than “impact score”, since the JIF is just one of many scores that can be used to assess a journal (and is known to be a methodologically deficient measure of mean citations).
I presented this study in the Greene Lab journal club, and we brainstormed a very interesting extension of Figure 5. Would it be possible to evaluate whether articles published behind a paywall received more preprint downloads following journal publication than those published open access? This could assess to what extent preprint servers were providing green open access to scholarly literature. It seems that this analysis would probably be best accomplished by integrating Unpaywall data as well as distinguishing bioRxiv downloads before and after journal publication. I am not sure whether the bioRxiv download stats are granular enough to support this temporal resolution? We found this potential analysis so interesting, we’ve decided to pursue it ourselves at https://github.com/greenelab/greenblack. While still a work in progress, feel free to get involved or investigate our results via that repository.
Figure 6 provides an interesting assessment of publishing delays by journal. There are many confounding factors, such as authors submitting to multiple journals before acceptance (authors may also post the preprint for convenience at journals that support direct bioRxiv submission) and preprints being posted at different stages of a study’s lifetime. Nonetheless, preprint timestamps provide an author-oriented measure of publishing delays, whereas previous largescale studies have used journal-reported timestamps. One criticism of journal reported timestamps is that journals try to minimize their publishing timeframes using practices such as “resetting the clock” upon each submitted version of a manuscript. Therefore, this study’s usage of a metric immune to journal manipulation is noteworthy. Figure 6 likely does truly reflect to some degree journals whose publication process is more cumbersome and slow, and hopefully this inspires some journals to optimize their workflows.
Data Availability
The study overall does an excellent job of making data and software available and documented. For example, I was able to submit a pull request to fix a broken link on rxivist.org. However, there is room for improvement.
I could not find the manuscript figures (or text) anywhere besides the PDF. Therefore, I had to take screenshots to extract the figures for my journal club presentation. It would be useful to add the figures to one of the existing repositories or data depositions.
The Zenodo archive with the data for the study and the Zenodo archive with the current database are currently released under a CC BY-NC 4.0 License. This license forbids commercial reuse and hence is not considered an open license. Furthermore, it is a poor choice for data. First, it is unclear whether copyright applies to any aspects of the created database in the United States. Therefore, some users may decide that either no copyright applies to the data or that their reuse is fair use. For these users, the CC license is irrelevant and can be ignored. However, more cautious users or those in other jurisdictions may feel restricted by the license and hence not use the data. The NC stipulation makes the data difficult to integrate with other data. For example, if copyright does apply, then the data would be incompatible with data licensed under CC BY-SA (share alike). Finally, attribution on a dataset level is often challenging, and social norms rather than legal recourse are generally sufficient. The authors should look into placing their datasets into the public domain via a CC0 waiver/license, which is becoming common and enables the information to be ingested by other data commons such as Wikidata. Finally, it is possible that users could rerun the open source code to regenerate the database, thereby creating a parellel version that is unencumbered by potential copyright issues.
The Rxivist API and dataset
It would be helpful to add a visualization of the database schema to the Methods to help readers quickly surmise the contents of the database. This schema should be automatically generated from the PostgreSQL database.
Conclusions
This study is a timely and intriguing analysis of bioRxiv preprints. Studies such as this are especially important to monitor how the transition to more open forms of scholarly publishing are progressing.
I do have one qualm with the study, not related to its content but instead to its effect. As the authors note, bioRxiv contains three times more preprints than the other servers indexed by PrePubMed combined. Rxivist adds to the network effect of bioRxiv by providing authors with additional visibility for their bioRxiv preprints. Therefore, it potentially has the effect of increasing bioRxiv’s dominance to the detriment of other preprint servers. My personal view is that the healthiest ecosystem for modernizing scholarly communication should have multiple preprint repositories to allow the most innovation and competition. Hopefully, preprint servers can begin to report similar data in standardized formats, such that resources like Rxivist can easily pull data from all compliant preprint servers.
Reviewer 2
In this paper, Abdil and Blekhman, present the first substantive analysis of submissions to bioRxiv, the leading preprint server in the life-sciences, and present a website, Rxivist, that collates information about Twitter activity and downloads per preprint.
Their analysis of the 35,000 odd preprints posted in the first five years of bioRxiv’s existence is thorough and much needed. At a superficial level it validates existing data from bioRxiv that shows a doubling-time of less than a year, culminating in the statistic that there were more preprints posted in 2018 than were posted in all the years preceding, combined. More interestingly, the authors present a field-by-field breakdown of this growth in pre-prints, showing a definite difference in their adoption in different life-science disciplines. Notably, neuroscientists and bioinformaticians have posted more preprints than any other group of researchers (Fig. 1a). This field-specific breakdown of preprint-adoption is important because it gives existing users within the field an idea of how much more preprint-advocacy is required within different fields. (One of my fields of research, plant biology, ranks very low on this list, something that hadn’t been evident to me previously.) I also hope that it guides the efforts of organisations like ASAPBio in targeting funding agencies (for e.g. in the non-biomedical life sciences) and groups in their advocacy efforts. One caveat in this analysis of course is that it doesn’t account for overall publishing volumes, which I expect vary quite a bit between fields.
This difference between publishing rates and cumulative publishing volumes is evident in Figure 2, where the authors present the readership statistics by field. This data, in my view, shows the interest level in preprints in different scientific communities, which perhaps also reflects broader community acceptance of preprints as a legitimate source of scientific research.
I also found the analysis of publication rates useful. The authors show that 67% of preprints posted between 2013 and 2016 were published in conventional peer-reviewed journals. Here again, their field-specific breakdown makes for interesting viewing, and pleasingly, it appears that the variation in publishing rates is much lower than the variation in number of submissions by field. This data is, again, like much of the rest of the paper, I think going to help make the case for preprints, though at least one commentator on social media has interpreted the 67% publication rate as “worrisome”.
The data on which journals have published the most preprints is also intriguing. The presence of megajournals like Scientific Reports and PLoS One high on this list is unsurprising, but it appears that Open Access journals (even selective ones like eLife and Nature Communications) seem to publish more preprints. More controversial though is the next analysis performed by the authors, i.e. the correlation between the Journal Impact Factor and preprint popularity (downloads).
This graph has received perhaps the most attention, even being reformatted and covered by Nature’s newsdesk. Personally, I find this analysis superfluous at best, and unwarranted at worst. The only utility I can see here is that it could perhaps guide authors to judge where to submit their research. The authors themselves make the point that this analysis is confounded by the fact that a lot of preprints could be downloaded after they are published in high IF journals. Another caveat is that preprint downloads are probably a reflection of the popularity of large labs, famous institutions, and famous PIs within the field, and this probably tracks with preprint acceptance at some of the highest IF journals. On the flip side, I suspect scientists with active social media accounts get more downloads of their preprints. I would also have liked a field specific breakdown of this correlation curve since I suspect fields like Neuroscience and Genomics (the more popular fields in terms of downloads) also routinely publish in higher IF journals compared to fields like Pharmacology & Toxicology, where preprints have less downloads. The point is, I suspect (like most journal-level metrics) this correlation is unlikely to help authors and readers much. On the other hand, this correlation might help convince preprint-sceptics, some of whom believe that bioRxiv will become a haven for bad research, that good science is and will be posted on bioRxiv.
While not discussed much in this preprint, the authors’ website, www.rxivist.org, features Twitter prominently, with the home-page featuring a list of the most tweeted about preprints in a 24 hour window. The site also has a “leaderboard” of authors with the most downloads and the preprints with the most downloads. I’m normally against the idea of leaderboards in science, but this list in particular could help trainees looking for labs with progressive ideas about publishing (since getting on the leaderboard effectively requires posting multiple preprints). I was also glad to note that the leaderboard itself appears to reflect a great diversity in terms of gender and geographical origin, something found in short supply in other ranked lists of scientists. In that sense I do hope the leaderboard receives some attention from journal editors soliciting reviewers and journalists looking for sources.
I found the website interesting to browse through when it first released, but I haven’t found myself visiting it again, or using it as a source for new preprints to read, and I’m sceptical that it will serve as such a tool. I typically find new preprints on my Twitter feed, and I find that to be a more personalised source of new preprints than sites like Rxivist, though obviously the website will be of much more value to scientists not on social media, who right now typically get preprints through word-of-mouth. Additionally, as preprint volumes grow, I do think services like Rxivist will gain more importance in searching for important advances within a field of research. I note that a similar sounding website (www.arxivist.com) exists for the arXiv (the preprint repository for the physical sciences), which provides individually customisable searches of preprints for users, and this could be one avenue of growth for Rxivist.
Rxivist may also be of great utility to science journalists looking for preprints to cover. I do however, think a live tally of the more global data that the authors present (number of downloads by field, submissions etc.) updated daily on the site would’ve been a useful feature to have on the website.
Overall, I found the analysis of bioRxiv preprints presented in the preprint very interesting, and of great value to preprint advocacy efforts. I can see the value of a resource like Rxivist and I also hope statistics like those shown there (downloads, tweets, altmetrics) would gain more traction in the current climate that’s focused on only citation-derived metrics like the IF and the h-index.
